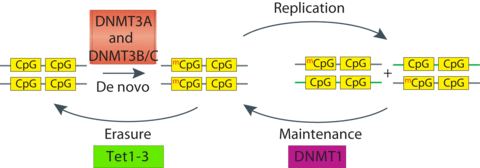
Mammalian DNA methylation
Our research program focuses on mechanistic understanding of protein machineries mediating key cellular activities, including epigenetic regulation. Epigenetics is a rapidly developing field that investigates how inheritable information beyond DNA sequence, such as chromatin modifications and non-coding RNAs, influences chromatin structure and function. Epigenetic modifications, including DNA methylation and histone modifications, regulate a broad spectrum of nuclear functions, including transcription, DNA replication and repair. Genomic patterns of DNA methylation and histone modifications are dynamically regulated by a set of chromatin modifiers, whose dysregulation is linked to many human diseases, such as cancer. We are particularly interested in mechanistic understanding of how DNA methylation machineries establish and maintain specific DNA methylation patterns, and how different histone modifications are recognized by distinct “reader” proteins to initiate downstream signaling.
Pathogen-Host Interaction
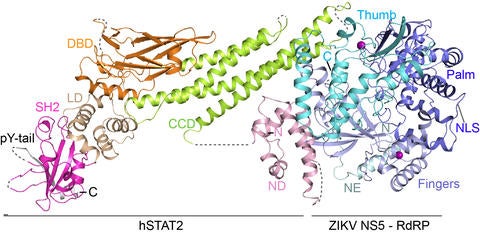
The battle between host and pathogen ultimately depends on the interaction between virulence proteins and host defense factors. We are interested in understanding how pathogen-host interactions influence viral infection and cellular activities, through structural, dynamic and functional characterizations of protein complexes that are important for pathogen infection or host immunity. Our work in the past has led to mechanistic elucidation of one important class of bacterial effector proteins, namely YopJ effectors (Zhang et al. NSMB 2016; Zhang et al. Nat. Plants 2017). In addition, we provided structural insights into the NS5 protein of Zika virus (ZIKV) (Wang et al. Nat. Commun. 2017), a virus-unique RNA-dependent RNA polymerase, as well as the molecular basis for the functional antagonism between ZIKV NS5 and human transcription factor STAT2 (Figure on right) (Wang et al. NSMB, 2020). Currently, we strive to determine the regulatory mechanism of viral replication machinery of SARS-CoV-2. By using X-ray crystallography and cryo-EM methods, combined with computational, biochemical and cellular analyses, we aim to elucidate the molecular mechanisms underlying these molecular regulations at atomic resolution.